BV 3D registration
- Make a transformation file for the epi data
- Registration between high and low resolution anatomical data
- Preprocessing :High resolution anatomical data
- Preprocessing :Low resolution anatomical data
- Talairach transformation (if necessary)
- Registration between the epi and high resolution anatomical data
Suppose we have the following three data.
- High resolution anatomical data (anat.high.vmr)
- Low resolution anatomical data (anat.low.vmr)
- EPI data (epi.fmr)
Make a transformation file for the epi data†
In this step, we will make a transformation file (.trf file). This .trf file adjusts the epi image to have the same directions as the high resolution anatomical image.
- Open the anat.high.vmr file
- In "3D Volume Tools" dialogue, choose "Coregistration" and click "Select FMR..." to open epi.fmr file for FMR-VMR coregistration.
Press "Align".
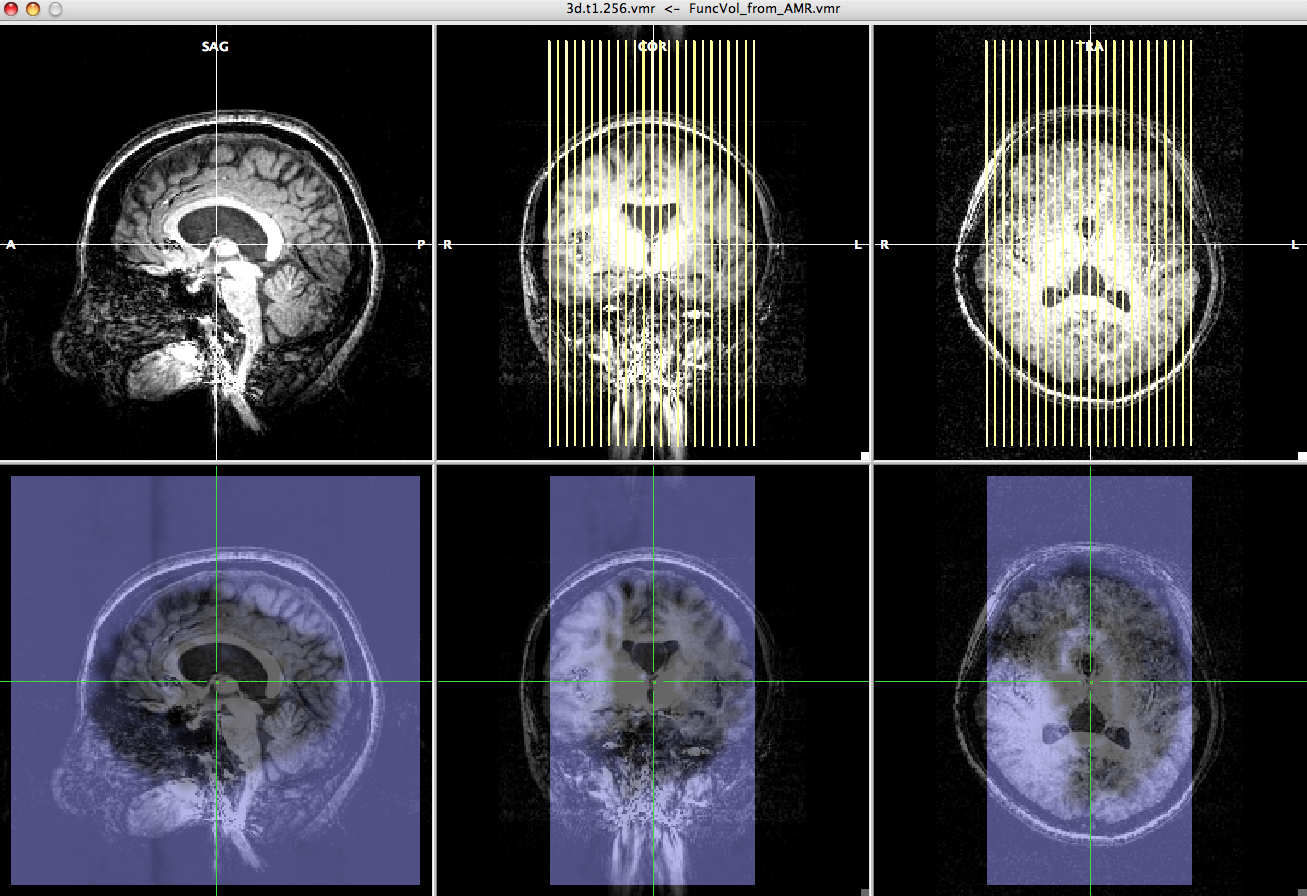
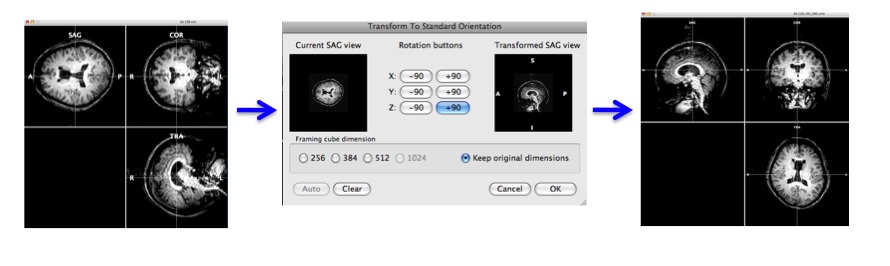
-> You will see "FMR-VMR Coregistration" dialogue, high resolution anatomical image and epi image. - Choose "Source Options" and click "To SAG".
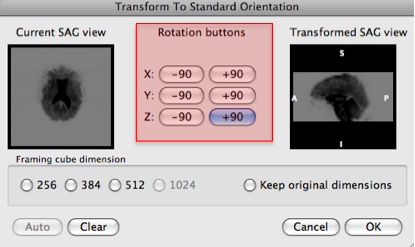
- Rotate the view by clicking the "Rotation buttons" to adjust the current sagital view to the sagital view of the anatomical data.
Press "OK".
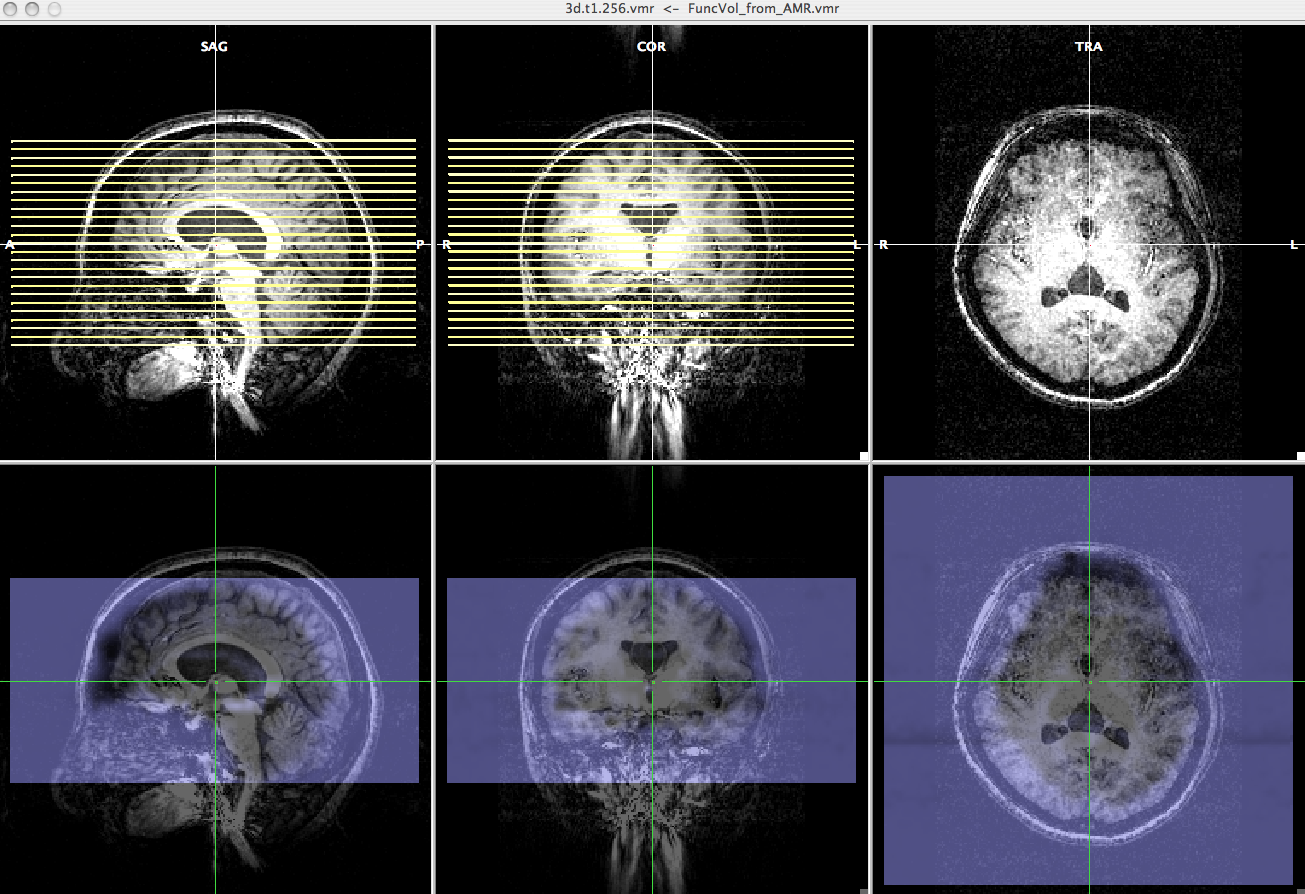
-> Now, you can check the epi image and high resolution anatomical image have roughly the same direction. - Select "Manual alignment..." for initial Alignment then press "Run IA". You will get a transformation file, epi-TO-anat.high_IA.trf.
※If there is a gap between the center of low resolution anatomical image and the center of epi image...
- Calculate the distance between these two centers (low resolution anatomical data and epi data) from pss value [cm].
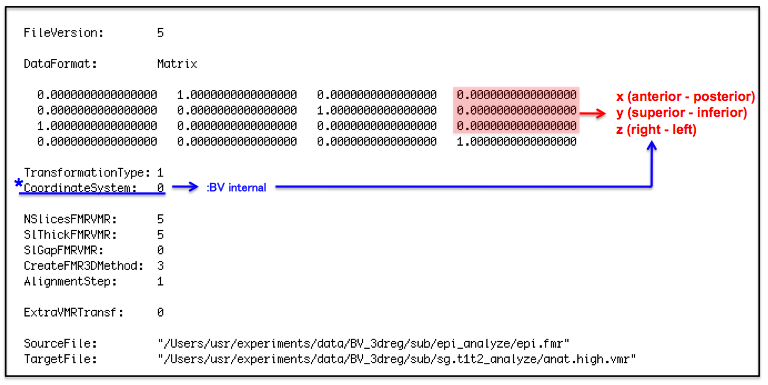
- Input the distance to the transformation matrix file, epi-TO-anat.high_IA.trf, in [mm].
↓epi-TO-anat.high_IA.trf file
*CoordinateSystem 0: BV internal [X: anterior -> posterior, Y: superior -> inferior, Z: right -> left] 1: BV-system [X: right -> left, Y: anterior -> posterior, Z: superior -> inferior] 2: Talairach [X: left -> right, Y: posterior -> anterior, Z: superior -> inferior]
Later on, use this new transformation matrix file as a epi-TO-anat.high_IA.trf file.
Registration between high and low resolution anatomical data†
Since the epi image has low resolution, we align the low resolution anatomical image instead of the epi image to high resolution anatomical image and make a transformation file. We will use this file for epi image - high resolution anatomical image coregistration.
Preprocessing :High resolution anatomical data†
- Open the anat.high.vmr file.
- In the "3D Volume Tools" dialogue, select "Talairach".
- Press "Find AC Point" and you will see "Find AC Point" dialogue. Move the cross to the AC point and click "OK".
- Press "Find AC-PC Plane..." Since the cross is located at AC, rotate the plane to find PC. When you find AC-PC plane, click OK".
- Press "Transform" and then click "GO".
-> You will get anat.high_ACPC.vmr and anat.high_ACPC.trf files.
Preprocessing :Low resolution anatomical data†
In this process, we will make the low resolution anatomical image to have the same view as the high resolution anatomical image for later alignment.
- Open anat.low.vmr file.
- Select "Spatial Transf" and click "To Sag" if the SAG view of the anat.low.vmr dataset does not show sagittal plane.
- Click "Iso-Voxel" and you will see "Iso-Voxel Transformation" dialogue. Check the Source voxel size is correct and press "OK".
-> You will get anat.low_SAG_ISO.vmr file.
- Open anat.high.vmr file
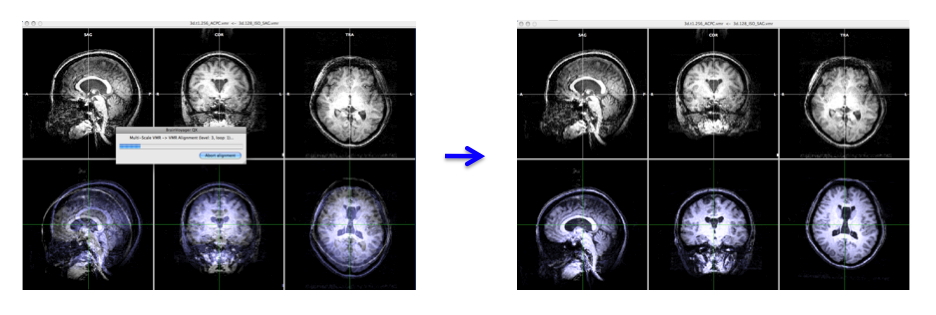
- In the "3D Volume Tools" dialogue, choose "Coregistration" and click "Select VMR..." button to select anat.low_SAG_ISO.vmr file for VMR-VMR coregistration.
Press "Align..." then "GO" in the "VMR-VMR Coregistration Options" dialogue. - Select "Spatial Transf" and click "Save .TRF" to save the parameter file for this spatial transformation.
-> You will get the anat.high_Man.trf file.
*Talairach transformation (if necessary)†
- #You can skip this part if you don't need the data in Talairach space.
- Open the anat.high_ACPC.vmr file.
- Select "Talairach" in the "3D Volume Tools".
- Click the list box in the "Talairach proportional grid reference points" and you will see eight reference points (AC, PC, AP, PP, SP, IP, RP, and LP).
- Select each point and move white cross to that point on the image to define the locations of all reference points.
Click the "Set point" each time when you define the point as the current location of the white cross. - After you define every reference points, click the "Save .TAL..." to save parameters. Click the "ACPC -> TAL..." then "Go".
-> Now you have anat.high_TAL.vmr and anat.high_ACPC.tal files.
Registration between the epi and high resolution anatomical data†
- Open the anat.high.vmr file.
# Open the anat.high_ACPC.vmr file if you want to check the image in Talairach space.
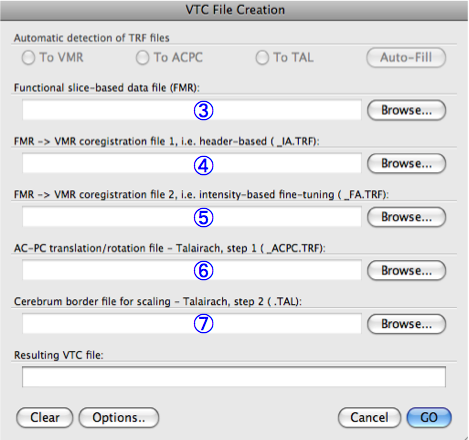
- Click "Analysis" and select "create 3D-Aligned Time course (VTC) data...".
- Click the "Browse..." to select the epi.fmr file for "Functional slice-based data file (FMR)"
- Select the identify matrix file, epi-TO-anat.high_IA.trf, for "FMR -> VMR coregistration file 1, i.e. header-based (_IA.TRF)".
- Select anat.high_Man.trf file for "FMR -> VMR coregistration file 2, i.e. intensity-based fine-tuning (_FA.TRF)".
Choose "yes" to the dialogue box asking "Do you really want to use it for FMR-TOVMR initial alignment?".
#You can skip the following two steps if you don't need the image in Talairach space.
- Select anat.high_ACPC.trf file for "AC-PC translation/rotation file - Talairach, step 1 (_ACPC.TRF)".
- Select anat.high_ACPC.tal file for "Cerebrum border file for scaling - Talairach, step 2 (.TAL)"
- Press "GO". When the process is done, you will get the epi.vtc file.
- Click the "Analysis" then select "Link 3D Time Course (VTC) File..."
- Press the "Browse" to choose the aligned file, epi.vtc and click "OK".
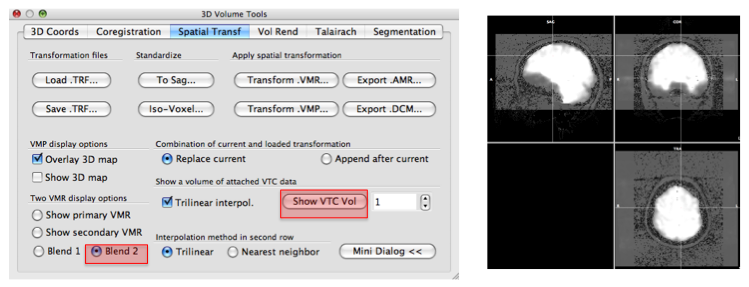
- On the "3D Volume Tools" dialogue, choose "Spatial Transf" and press "Show VTC Vol" and after a while, you will see the image aligned epi image.
- Click the "Blend2" and you can check if the epi image are correctly aligned to anatomy image.
Attach file: